2024
Cacioppo, R.*, Gillis, A.*, Shlamovitz, I.*, Zeller, A.*, Castiblanco, D., Crisp, A., Haworth, B., Arabiotorre, A., Abyaneh, P., Bao, Y., Sale, J.E., Berry, S.^, Tufegdžić Vidaković, A.^
Molecular Cell
| PDF
| Journal
| UNSW news
| MRC news
^ Corresponding authors
Arrayed CRISPR libraries for the genome-wide activation, deletion and silencing of human protein-coding genes
Yin, J.-A., Frick, L., Scheidmann, M. C., Liu, T., Trevisan, C., Dhingra, A., Spinelli, A., Wu, Y., Yao, L., Vena, D. L., Knapp, B., Guo, J., Cecco, E. D., Ging, K., Armani, A., Oakeley, E. J., Nigsch, F., Jenzer, J., Haegele, J., Pikusa, M., Täger, J., Rodriguez-Nieto, S., Bouris, V., Ribeiro, R., Baroni, F., Bedi, M. S., Berry, S., Losa, M., Hornemann, S., Kampmann, M., Pelkmans, L., Hoepfner, D., Heutink, P. & Aguzzi, A.
Nat. Biomed. Eng.
| PDF
| Journal
| bioRxiv
Global control of RNA polymerase II
Gillis, A., and Berry, S.
BBA Gene Regulatory Mechanisms
| PDF
| Journal
2023
Integrating analog and digital modes of gene expression at Arabidopsis FLC
Antoniou-Kourounioti, R.L.*, Meschichi, A.*, Reeck, S.*, Berry, S., Menon, G., Zhao, Y., Fozard, J., Holmes, T., Zhao, L., Wang, H., Hartley, M., Dean, C., Rosa, S., Howard, M.
eLife
| PDF
| Journal
| bioRxiv
* Equal contribution
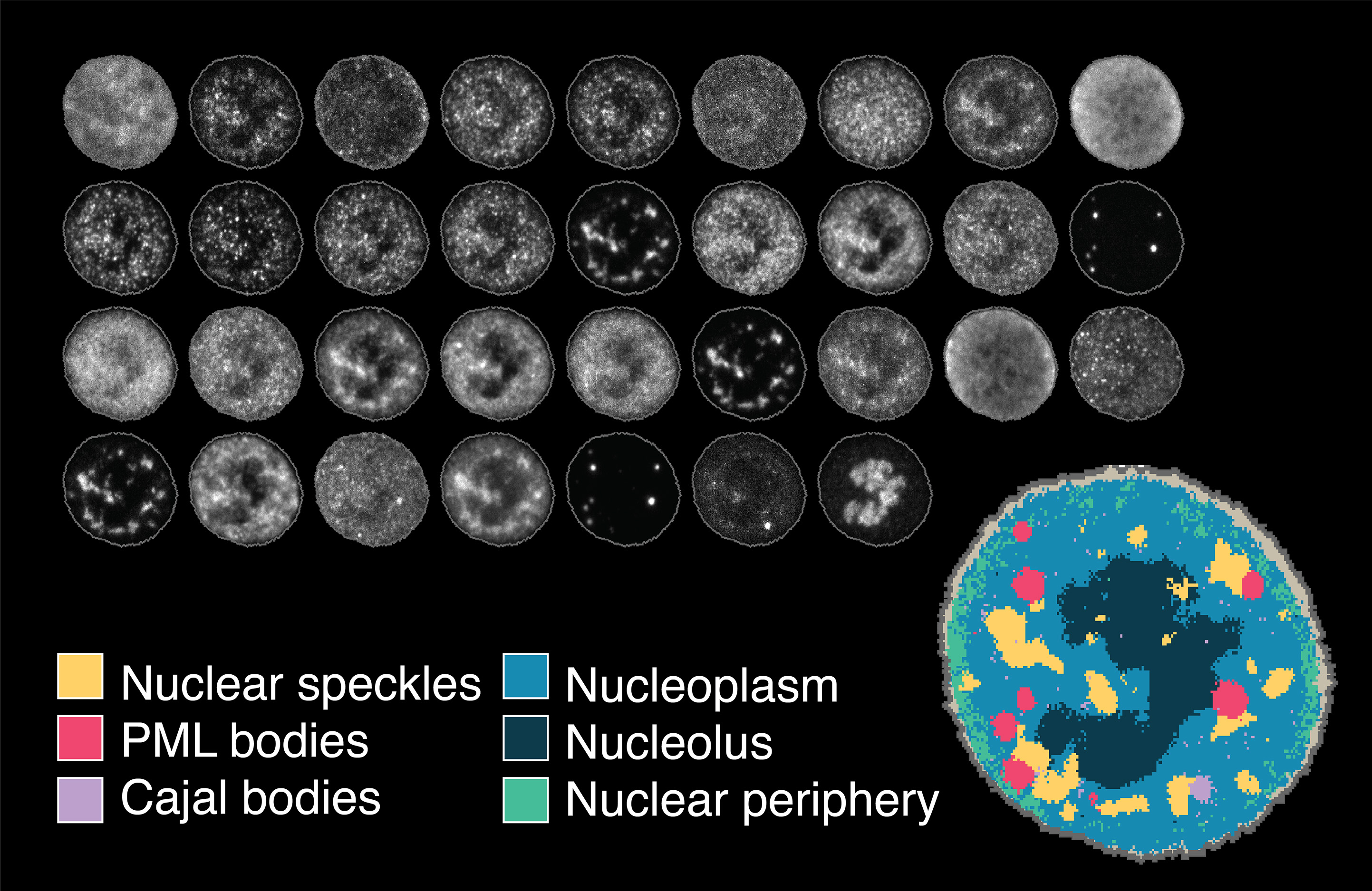
Learning consistent subcellular landmarks to quantify changes in multiplexed protein maps
Spitzer, H.*, Berry, S.*, Donoghoe, M., Pelkmans, L. and Theis, F.
Nature Methods
| PDF
| Journal
| Research Briefing
| UNSW news
| bioRxiv
| GitHub
| Read the Docs
* Equal contribution
2022
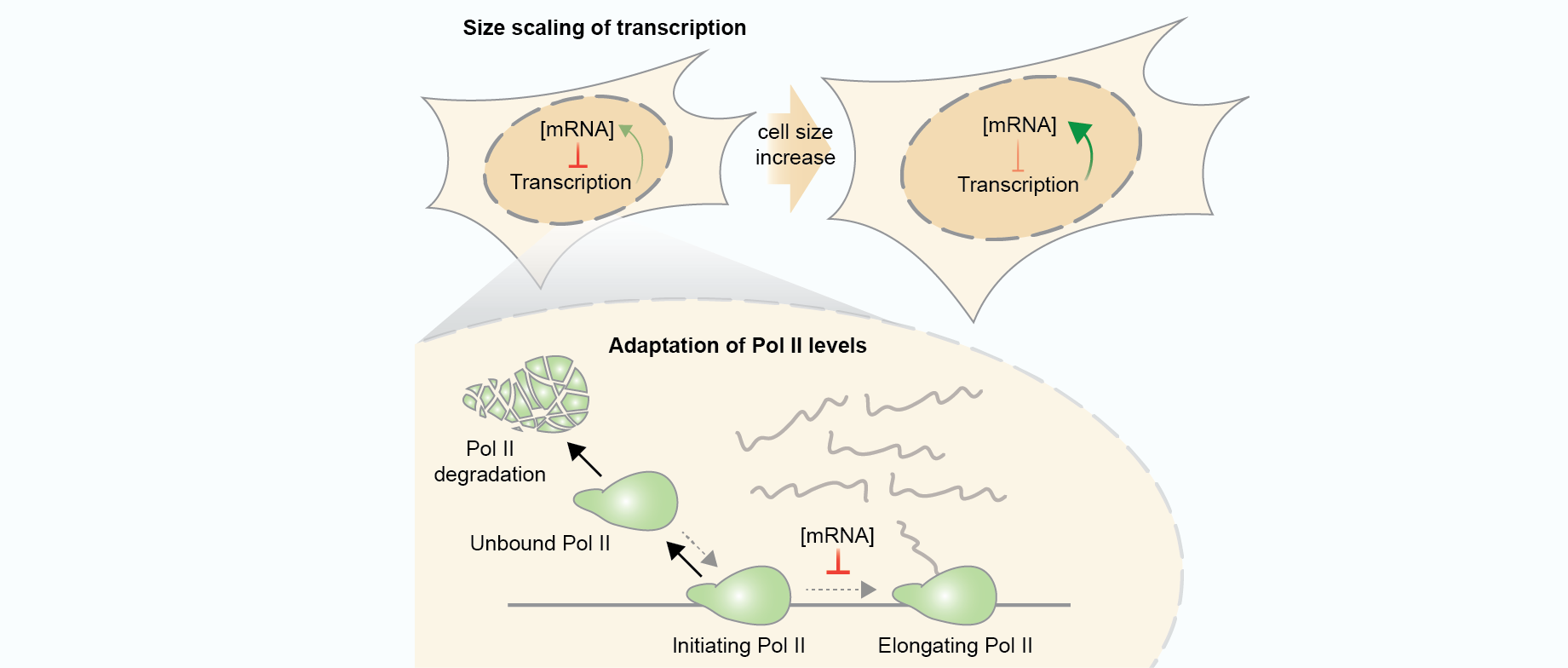
Mechanisms of cellular mRNA transcript homeostasis
Berry, S., and Pelkmans, L.
Trends in Cell Biology
| Journal
Berry, S.^, Müller, M., Rai, A. and Pelkmans, L.^
Cell Systems
| PDF
| Journal
| bioRxiv
^ Corresponding authors
2021
High content genome-wide siRNA screen to investigate the coordination of cell size and RNA production
Müller, M., Avar, M., Heinzer, D., Emmenegger, M., Aguzzi, A., Pelkmans, L.^, and Berry, S.^
Scientific Data
| PDF
| Journal
| Data
^ Corresponding authors
2017
Berry, S., Rosa, S., Howard, M., Bühler, M., and Dean, C.
Genes & Development
| PDF
| Journal
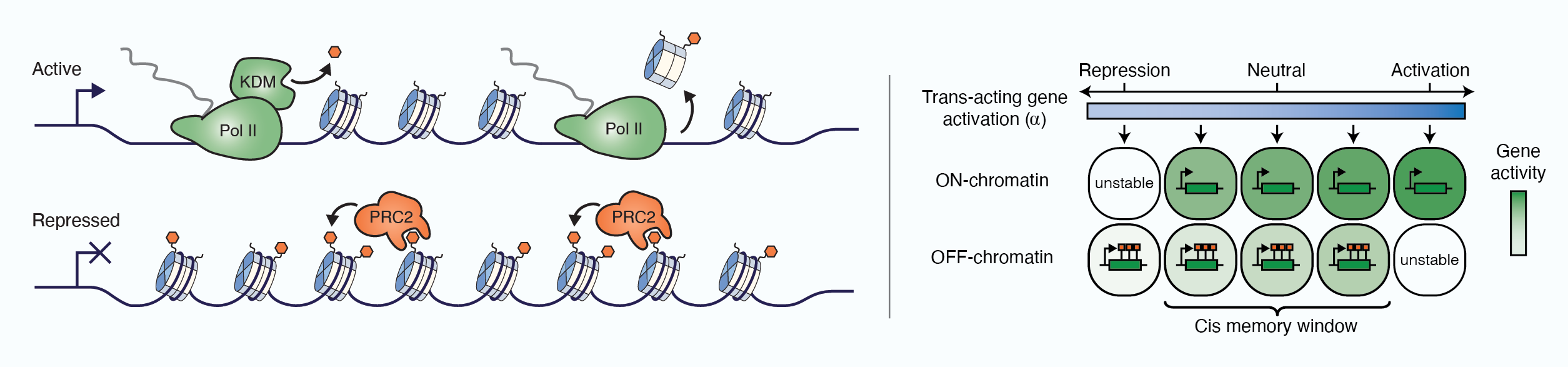
Slow Chromatin Dynamics Allow Polycomb Target Genes to Filter Fluctuations in Transcription Factor Activity
Berry, S., Dean, C., and Howard, M.
Cell Systems
| PDF
| Supplementary
| Journal
| Commentary
Distinct phases of Polycomb silencing to hold epigenetic memory of cold in Arabidopsis
Yang, H.*, Berry, S.*, Olsson, T.S.G., Hartley, M., Howard, M., and Dean, C.
Science
| PDF (accepted version)
| Journal
* Equal contribution
2015
Local chromatin environment of a Polycomb target gene instructs its own epigenetic inheritance
Berry, S., Hartley, M., Olsson, T.S.G., Dean, C., and Howard, M.
eLife
| PDF
| Journal
Environmental perception and epigenetic memory: mechanistic insight through FLC
Berry, S., and Dean, C.
The Plant Journal
| PDF
| Journal
2014
Lipopolysaccharide O antigen size distribution is determined by a chain extension complex of variable stoichiometry in Escherichia coli O9a
King, J.D.*, Berry, S.*, Clarke, B.R., Morris, R.J., and Whitfield, C.
Proceedings of the National Academy of Sciences
| PDF
| Supplementary
| Journal
* Equal contribution
2012
Illustrations of Mathematical Modeling in Biology: Epigenetics, Meiosis, and an Outlook
Richards, D.*, Berry, S.*, and Howard, M.
Cold Spring Harbor Symposia on Quantitative Biology
| PDF
| Journal
* Equal contribution
2011
qwViz: Visualisation of quantum walks on graphs
Berry, S.D., Bourke, P., and Wang, J.B.
Computer Physics Communications
| Journal
Two-particle quantum walks: Entanglement and graph isomorphism testing
Berry, S.D., and Wang, J.B.
Physical Review A
| Journal
2010
Quantum-walk-based search and centrality
Berry, S.D., and Wang, J.B.
Physical Review A
| arXiv
| Journal








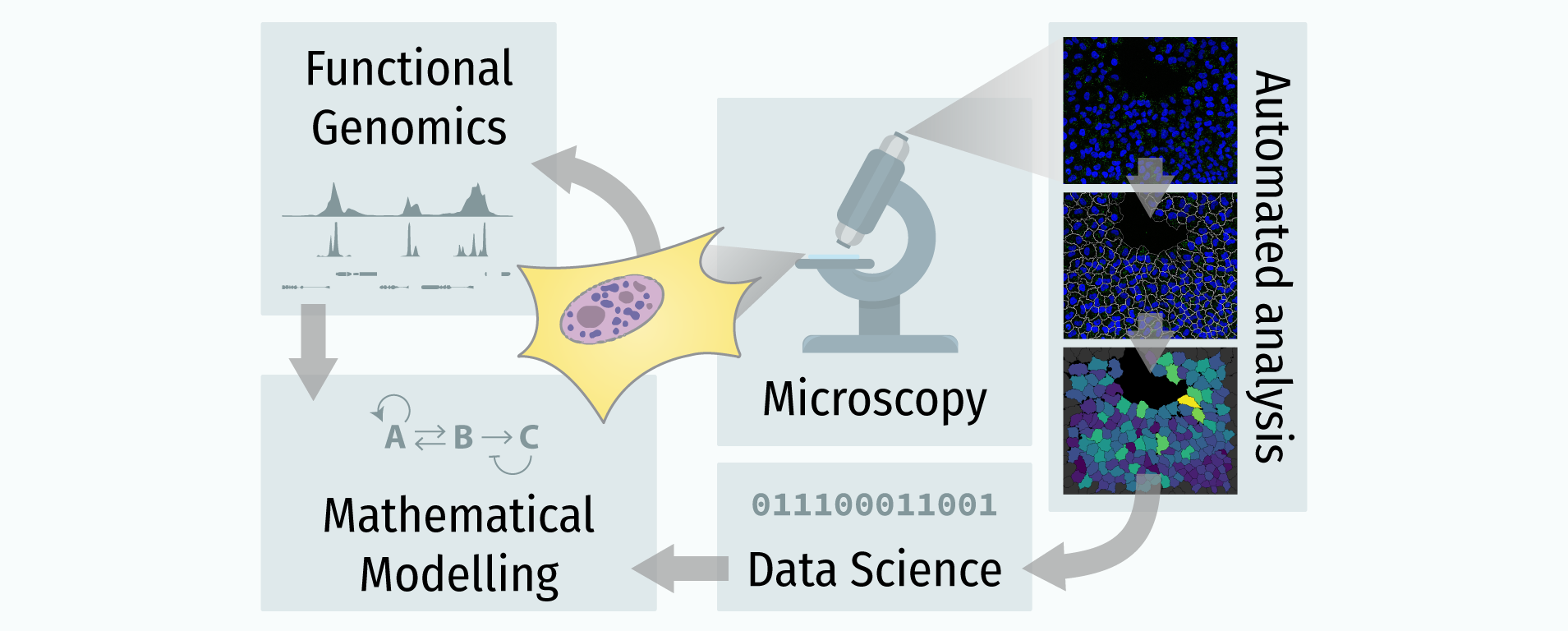
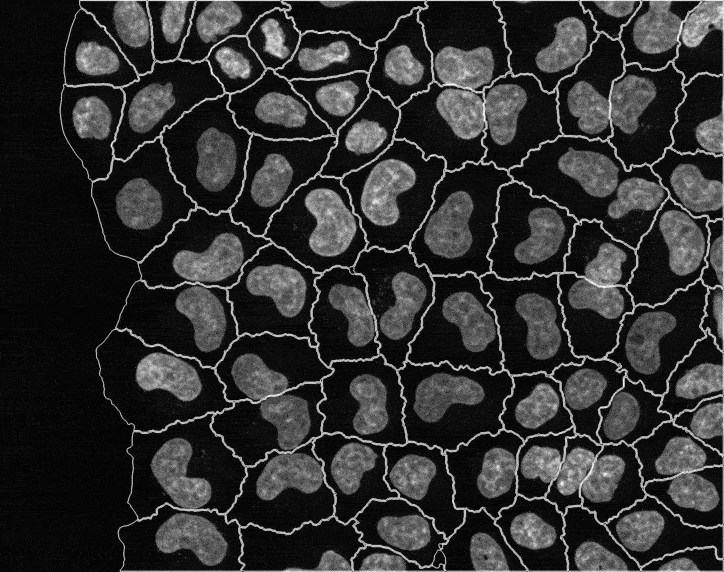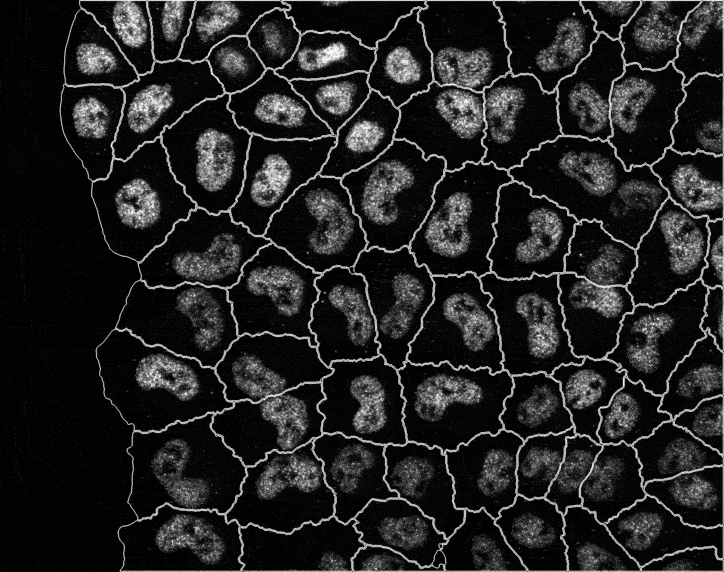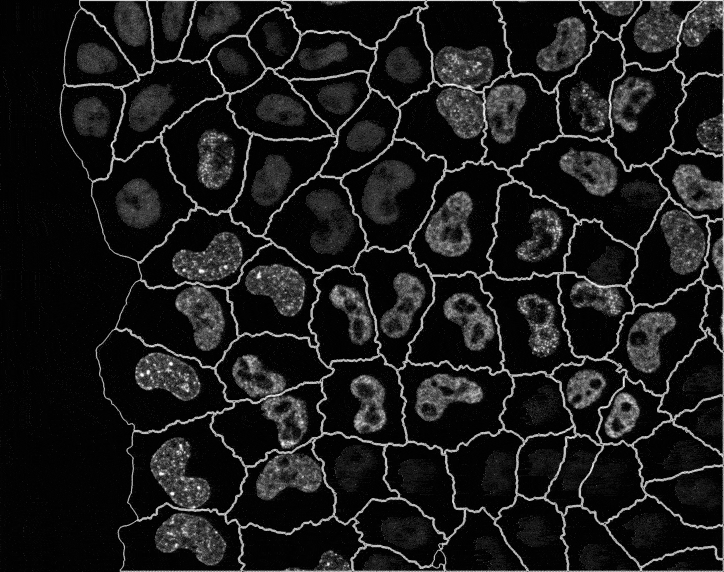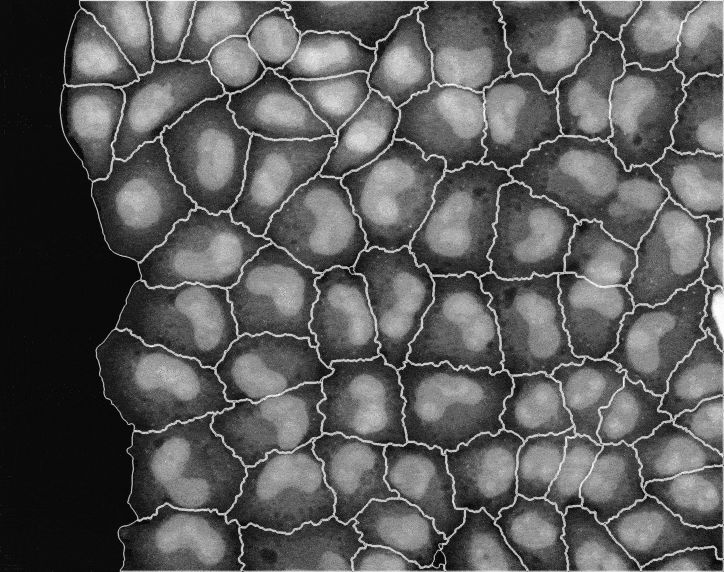 In recent years, technological advances have made it possible to visualise tens to hundreds of proteins in single cells. This has brought with it the ability to accurately quantify the high-dimensional relationships between protein abundances and their modification states, as well as the cell-to-cell variability of their subcellular localisation and co-localisation.
In recent years, technological advances have made it possible to visualise tens to hundreds of proteins in single cells. This has brought with it the ability to accurately quantify the high-dimensional relationships between protein abundances and their modification states, as well as the cell-to-cell variability of their subcellular localisation and co-localisation.